AI makes huge progress predicting how proteins fold – one of biology's greatest challenges – promising rapid drug development
- Written by Marc Zimmer, Professor of Chemistry, Connecticut College
Takeaways
A “deep learning” software program from Google-owned lab DeepMind showed great progress in solving one of biology’s greatest challenges – understanding protein folding.
Protein folding is the process by which a protein takes its shape from a string of building blocks to its final three-dimensional structure, which determines its function.
By better predicting how proteins take their structure, or “fold,” scientists can more quickly develop drugs that, for example, block the action of crucial viral proteins.
Solving what biologists call “the protein-folding problem” is a big deal. Proteins are the workhorses of cells and are present in all living organisms. They are made up of long chains of amino acids and are vital for the structure of cells and communication between them as well as regulating all of the chemistry in the body.
This week, the Google-owned artificial intelligence company DeepMind[1] demonstrated a deep-learning program called AlphaFold2[2], which experts are calling a breakthrough[3] toward solving the grand challenge of protein folding[4].
Proteins are long chains of amino acids linked together like beads on a string. But for a protein to do its job in the cell, it must “fold” – a process of twisting and bending that transforms the molecule into a complex three-dimensional structure that can interact with its target in the cell. If the folding is disrupted, then the protein won’t form the correct shape – and it won’t be able to perform its job inside the body. This can lead to disease – as is the case in a common disease like Alzheimer’s, and rare ones like cystic fibrosis.
Deep learning is a computational technique that uses the often hidden information contained in vast datasets to solve questions of interest. It’s been used widely in fields such as games, speech and voice recognition, autonomous cars, science and medicine.
I believe that tools like AlphaFold2 will help scientists to design new types of proteins, ones that may, for example, help break down plastics and fight future viral pandemics and disease.
I am a computational chemist[5] and author of the book The State of Science[6]. My students and I study the structure and properties of fluorescent proteins[7] using protein-folding computer programs based on classical physics.
After decades of study by thousands of research groups, these protein-folding prediction programs are very good at calculating structural changes that occur when we make small alterations to known molecules.
But they haven’t adequately managed to predict how proteins fold from scratch. Before deep learning came along, the protein-folding problem seemed impossibly hard, and it seemed poised to frustrate computational chemists for many decades to come.
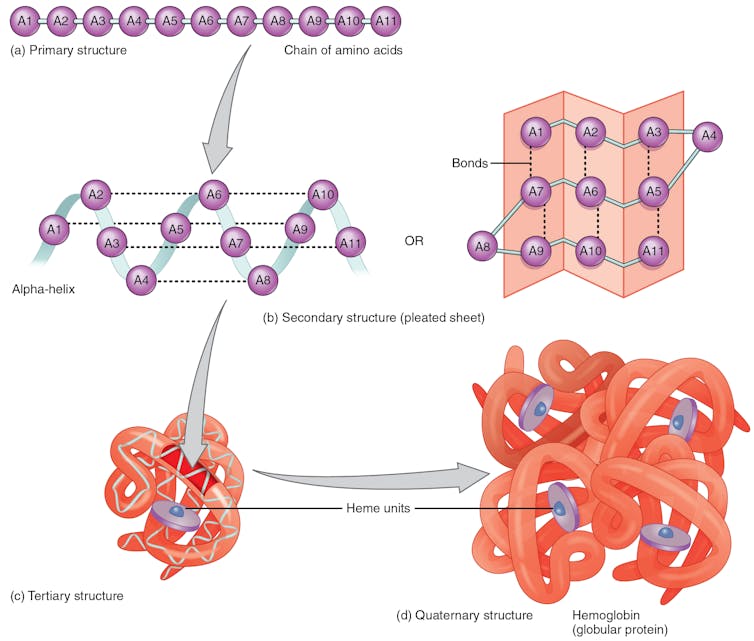 A chain of amino acids goes through several folding steps, which occurs through hydrogen bonds between amino acids in different regions of the protein, before arriving at the final structure. The example shown here is hemoglobin, a protein in red blood cells that transports oxygen to body tissues.
Anatomy & Physiology, Connexions website, CC BY[8][9]
A chain of amino acids goes through several folding steps, which occurs through hydrogen bonds between amino acids in different regions of the protein, before arriving at the final structure. The example shown here is hemoglobin, a protein in red blood cells that transports oxygen to body tissues.
Anatomy & Physiology, Connexions website, CC BY[8][9]
Protein folding
The sequence of the amino acids – which is encoded in DNA – defines the protein’s 3D shape. The shape determines its function. If the structure of the protein changes, it is unable to perform its function. Correctly predicting protein folds based on the amino acid sequence could revolutionize drug design, and explain the causes of new and old diseases.
All proteins with the same sequence of amino acid building blocks fold into the same three-dimensional form, which optimizes the interactions between the amino acids. They do this within milliseconds, although they have an astronomical number of possible configurations available to them – about 10 to the power of 300[10]. This massive number is what makes it hard to predict how a protein folds even when scientists know the full sequence of amino acids that go into making it. Previously predicting the structure of protein from the amino acid sequence was impossible. Protein structures were experimentally determined, a time-consuming and expensive endeavor.
Once researchers can better predict how proteins fold, they’ll be able to better understand how cells function and how misfolded proteins cause disease. Better protein prediction tools will also help us design drugs that can target a particular topological region of a protein where chemical reactions take place.
 What’s your move?
style-photography/Getty Images[11]
What’s your move?
style-photography/Getty Images[11]
AlphaFold is born from deep-learning chess, Go and poker games
The success of DeepMind’s protein-folding prediction program, called AlphaFold[12], is not unexpected. Other deep-learning programs written by DeepMind[13] have demolished the world’s best chess, Go and poker players.
In 2016 Stockfish-8[14], an open-source chess engine, was the world’s computer chess champion. It evaluated 70 million chess positions per second and had centuries of accumulated human chess strategies and decades of computer experience to draw upon. It played efficiently and brutally, mercilessly beating all its human challengers without an ounce of finesse. Enter deep learning.
On Dec. 7, 2017, Google’s deep-learning chess program AlphaZero[15] thrashed Stockfish-8. The chess engines played 100 games, with AlphaZero winning 28 and tying 72. It didn’t lose a single game. AlphaZero did only 80,000 calculations per second, as opposed to Stockfish-8’s 70 million calculations, and it took just four hours to learn chess from scratch by playing against itself a few million times and optimizing its neural networks as it learned from its experience.
AlphaZero[16] didn’t learn anything from humans or chess games played by humans. It taught itself and, in the process, derived strategies never seen before. In a commentary[17] in Science magazine, former world chess champion Garry Kasparov wrote that by learning from playing itself, AlphaZero developed strategies that “reflect the truth” of chess rather than reflecting “the priorities and prejudices” of the programmers. “It’s the embodiment of the cliché ‘work smarter, not harder.’”
How do proteins fold?CASP – the Olympics for molecular modelers
Every two years, the world’s top computational chemists test the abilities of their programs to predict the folding of proteins and compete in the Critical Assessment of Structure Prediction[18] (CASP) competition.
In the competition, teams are given the linear sequence of amino acids for about 100 proteins for which the 3D shape is known but hasn’t yet been published; they then have to compute how these sequences would fold. In 2018 AlphaFold, the deep-learning rookie at the competition, beat all the traditional programs – but barely.
Two years later, on Monday, it was announced that Alphafold2 had won the 2020 competition by a healthy margin. It whipped its competitors, and its predictions were comparable to the existing experimental results determined through gold standard techniques like X-ray diffraction crystallography and cryo-electron microscopy. Soon I expect AlphaFold2 and its progeny will be the methods of choice to determine protein structures before resorting to experimental techniques that require painstaking, laborious work on expensive instrumentation.
One of the reasons for AlphaFold2’s success is that it could use the Protein Database[19], which has over 170,000 experimentally determined 3D structures, to train itself to calculate the correctly folded structures of proteins.
The potential impact of AlphaFold can be appreciated if one compares the number of all published protein structures – approximately 170,000 – with the 180 million DNA and protein sequences deposited in the Universal Protein Database[20]. AlphaFold will help us sort through treasure troves of DNA sequences hunting for new proteins with unique structures and functions[21].
Has AlphaFold made me, a molecular modeler, redundant?
As with the chess and Go programs – AlphaZero and AlphaGo – we don’t exactly know what the AlphaFold2 algorithm is doing and why it uses certain correlations, but we do know that it works.
Besides helping us predict the structures of important proteins, understanding AlphaFold’s “thinking” will also help us gain new insights into the mechanism of protein folding.
[Deep knowledge, daily. Sign up for The Conversation’s newsletter[22].]
One of the most common fears expressed about AI is that it will lead to large-scale unemployment. AlphaFold still has a significant way to go before it can consistently and successfully predict protein folding.
However, once it has matured and the program can simulate protein folding, computational chemists will be integrally involved in improving the programs, trying to understand the underlying correlations used, and applying the program to solve important problems such as the protein misfolding associated with many diseases such as Alzheimer’s, Parkinson’s, cystic fibrosis and Huntington’s disease.
AlphaFold and its offspring will certainly change the way computational chemists work, but it won’t make them redundant. Other areas won’t be as fortunate. In the past robots were able to replace humans doing manual labor; with AI, our cognitive skills are also being challenged.
References
- ^ DeepMind (www.deepmind.com)
- ^ AlphaFold2 (deepmind.com)
- ^ breakthrough (www.nature.com)
- ^ protein folding (doi.org)
- ^ I am a computational chemist (scholar.google.com)
- ^ The State of Science (rowman.com)
- ^ fluorescent proteins (www.conncoll.edu)
- ^ Anatomy & Physiology, Connexions website (upload.wikimedia.org)
- ^ CC BY (creativecommons.org)
- ^ about 10 to the power of 300 (web.archive.org)
- ^ style-photography/Getty Images (www.gettyimages.com)
- ^ AlphaFold (deepmind.com)
- ^ DeepMind (deepmind.com)
- ^ Stockfish-8 (www.chessprogramming.org)
- ^ AlphaZero (doi.org)
- ^ AlphaZero (web.stanford.edu)
- ^ commentary (doi.org)
- ^ Critical Assessment of Structure Prediction (predictioncenter.org)
- ^ Protein Database (www.rcsb.org)
- ^ Universal Protein Database (www.uniprot.org)
- ^ functions (deepmind.com)
- ^ Sign up for The Conversation’s newsletter (theconversation.com)
Authors: Marc Zimmer, Professor of Chemistry, Connecticut College