Scientists at work: We use environmental DNA to monitor how human activities affect life in rivers and streams
- Written by Marie Simonin, Research Scientist, Inrae
Rivers, lakes and wetlands cover just 1% of the Earth’s surface but are home to nearly 10% of all species, including fish, mammals, birds, insects and crustaceans. But these rich, diverse[1] ecosystems are in free fall[2]. Worldwide, species are declining faster now[3] than at any other time in human history, and fresh waters are losing more species than land or ocean ecosystems.
Today about 1 in 4 freshwater creatures face extinction[4]. Wetlands are disappearing three times faster than forests[5]. Across the globe, water quality is plummeting, polluted by plastic, sewage, mining sludge, industrial and agricultural chemicals and much more[6].
It’s challenging to study how these stresses are affecting aquatic life. There are many diverse threats, and river networks cover broad geographic regions. Often they run through remote, nearly inaccessible areas. Current techniques for monitoring freshwater species are labor-intensive and costly[7].
In our work[8] as researchers[9] in ecology[10], we are testing a new method that can vastly expand biomonitoring: using environmental DNA, or eDNA, in rivers to catalog and count species[11]. Federal and local agencies need this data to restore water quality and save dwindling species from extinction.
This preview of the film “Hidden Rivers” reveals the diverse and little-known life in Southern Appalachian waterways.Traditional methods are slow and expensive
With traditional biomonitoring methods, scientists count individual species and their abundance at just a few sites[12]. For example, one recent study of mountaintop mining impacts on fish in West Virginia[13] sampled just four sites with a team of four researchers.
Collecting and identifying aquatic organisms requires highly skilled ecologists and taxonomists with expertise in a wide variety of freshwater species. For each sample of fish or invertebrates collected in the field, it takes from hours to weeks to identify all of the species. Only wealthy nations can afford this costly process.
Conserving threatened and endangered species and keeping river ecosystems healthy requires monitoring broad areas over time. Sensitive aquatic insects and fish species are the freshwater equivalent of the proverbial canary in a coal mine: If these species are absent, that’s a strong indicator of water quality problems. The cause may be mining, agriculture, urbanization or other sources, as well as dams[14] that block animals’ downstream movements.
Scientists sample for fish in a Maryland stream by ‘electrofishing’ – stunning fish with a mild electrical pulse so they can be collected, identified and released after the shock wears off.Free-floating genetic evidence
Innovations in genetic technology have created a powerful, affordable new tool that we are now testing. The process involves extracting eDNA from genetic material floating in the water – skin, scales, feces and single-celled organisms, such as bacteria.
By analyzing this genetic information, we can detect a wide range of species[15]. We started considering using eDNA for our research in 2018, after several studies demonstrated its power to monitor single species of interest or groups of organisms in rivers[16] and oceans[17].
Collecting eDNA is easy: One 4-ounce water sample can capture remnant DNA from thousands of aquatic species. Another benefit is that it doesn’t require killing wildlife for identification.
In the lab, we analyze the DNA from different taxonomic groups one by one: bacteria, algae, fish and macroinvertebrates[18] – organisms that lack backbones and are large enough to see, such as snails, worms and beetles. Many researchers study just one group, but we assess all of them at the same time.
We then match our DNA sequences with freshwater species that are already catalogued in existing databases. In this way, we can chart the distribution and abundance of these organisms within and across rivers.
This process requires just a cheap filter, a syringe and vials, and anyone can do it. Commercial eDNA companies charge less than $200 to extract and sequence a sample.
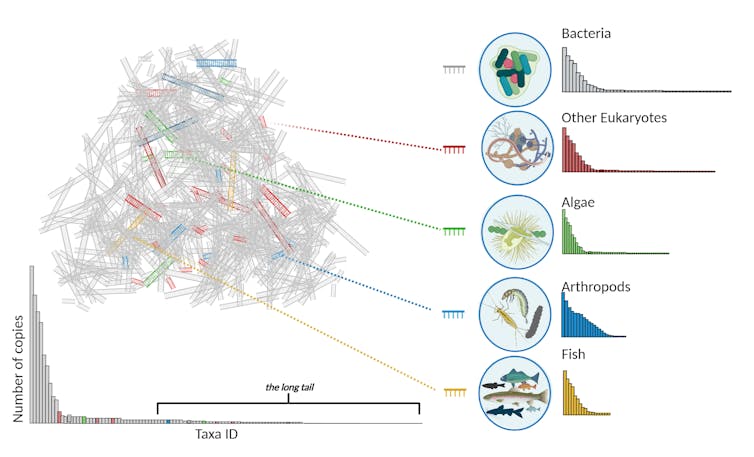 Most eDNA the authors collect from streams is microbial (the gray DNA in the cartoon above). Without special techniques, they would not ‘see’ the less frequent DNA from other taxonomic groups, so their surveys would generate a species abundance curve like the one on the bottom left, in which most groups of conservation concern are too rare to detect or fall into the ‘long tail’ of rare occurrences. By using targeted primers – short stretches of DNA that are unique to specific groups of organisms – they can amplify the eDNA of less abundant groups, like algae, arthropods and fish, as shown on the right.
Emily Bernhardt, produced using Biorender, CC BY-ND[19]
Most eDNA the authors collect from streams is microbial (the gray DNA in the cartoon above). Without special techniques, they would not ‘see’ the less frequent DNA from other taxonomic groups, so their surveys would generate a species abundance curve like the one on the bottom left, in which most groups of conservation concern are too rare to detect or fall into the ‘long tail’ of rare occurrences. By using targeted primers – short stretches of DNA that are unique to specific groups of organisms – they can amplify the eDNA of less abundant groups, like algae, arthropods and fish, as shown on the right.
Emily Bernhardt, produced using Biorender, CC BY-ND[19]
Altered rivers
Using this method, we extensively surveyed 93 rivers in West Virginia – looking at the entire tree of life, from the tiniest bacteria to fish – in two days with a four-person team.
The Appalachian rivers that we study teem with life. These are some of the world’s most biologically diverse temperate freshwater ecosystems, home to many fish species[20], as well as salamanders, crayfish, mussels and aquatic insects. Many are found nowhere else[21]. We tallied more than 10,000 different species[22] in those 93 waterways.
The area where we worked is an intensive coal mining region, which heavily affects waterways. Liquids draining from mines are acidic[23], but in this region they react with limestone rock, so the net effect is to make local streams alkaline. Mine drainage also increases streams’ salinity and concentrations of sulfate and other contaminants[24]. Our research revealed that mined watersheds held 40% fewer species than areas without mining operations, and the organisms we detected were less abundant than in unaffected rivers.
Assessing river health
We believe this new approach represents a revolution for biomonitoring, expanding our ability to quantify and study freshwater life. It’s also an important new conservation tool, allowing scientists to track changes in populations of endangered or invasive species. Researchers also can use eDNA to monitor biodiversity or discover new species in oceans or soils.
This open-science method[25] makes all DNA data widely available, with nearly all sequences placed in public repositories. Moving forward, we expect that it will aid many types of research, as well as state and local monitoring and conservation programs. Investments in collecting eDNA and identifying organisms and analyzing their genetic signatures will continue to make it a more effective tool.
[Over 100,000 readers rely on The Conversation’s newsletter to understand the world. Sign up today[26].]
Efforts are underway to better target various individual species, focusing on those that are endangered, invasives that damage ecosystems and sensitive species that serve as indicators of river health. Scientists are freezing eDNA samples at -112 degrees F (-80 C) in expectation that technological advances may yield more information in the future[27].
Traditional monitoring approaches remain valuable, but eDNA adds an important new tool to the toolkit. Together, these approaches can begin to answer many questions about food webs, the conservation status of species, reproduction rates, species interactions, organisms’ health, disease and more.
References
- ^ diverse (wwfcee.org)
- ^ in free fall (wwf.panda.org)
- ^ declining faster now (www.un.org)
- ^ 1 in 4 freshwater creatures face extinction (www.iucn.org)
- ^ three times faster than forests (doi.org)
- ^ plastic, sewage, mining sludge, industrial and agricultural chemicals and much more (uneplive.unep.org)
- ^ labor-intensive and costly (doi.org)
- ^ work (esajournals.onlinelibrary.wiley.com)
- ^ researchers (scholar.google.fr)
- ^ ecology (scholar.google.com)
- ^ catalog and count species (doi.org)
- ^ a few sites (doi.org)
- ^ mountaintop mining impacts on fish in West Virginia (doi.org)
- ^ dams (www.nwf.org)
- ^ detect a wide range of species (doi.org)
- ^ rivers (doi.org)
- ^ oceans (theconversation.com)
- ^ macroinvertebrates (www.epa.gov)
- ^ CC BY-ND (creativecommons.org)
- ^ many fish species (www.conservationfisheries.org)
- ^ Many are found nowhere else (www.natureserve.org)
- ^ more than 10,000 different species (doi.org)
- ^ acidic (www.epa.gov)
- ^ sulfate and other contaminants (doi.org)
- ^ open-science method (doi.org)
- ^ Sign up today (theconversation.com)
- ^ more information in the future (doi.org)
Authors: Marie Simonin, Research Scientist, Inrae