Mixed-ancestry genetic research shows a bit of Native American DNA could reduce risk of Alzheimer's disease
- Written by Diane Xue, Public Health Genetics PhD student and Research Assistant in Medical Genetics, University of Washington
Since the human genome was first mapped, scientists have discovered hundreds of genes influencing illnesses like breast cancer, heart disease and Alzheimer’s disease. Unfortunately, Black people, Indigenous people and other people of color are underrepresented in most genetic studies[1]. This has resulted in a skewed and incomplete understanding[2] of the genetics of many diseases.
We are two researchers who have been working to find genes[3] that affect people’s risk[4] for various diseases. Our team recently found a genetic region that appears to be protective against Alzheimer’s disease[5]. To do this, we used a method called admixture mapping that uses data from people with mixed ancestry to find genetic causes of disease.
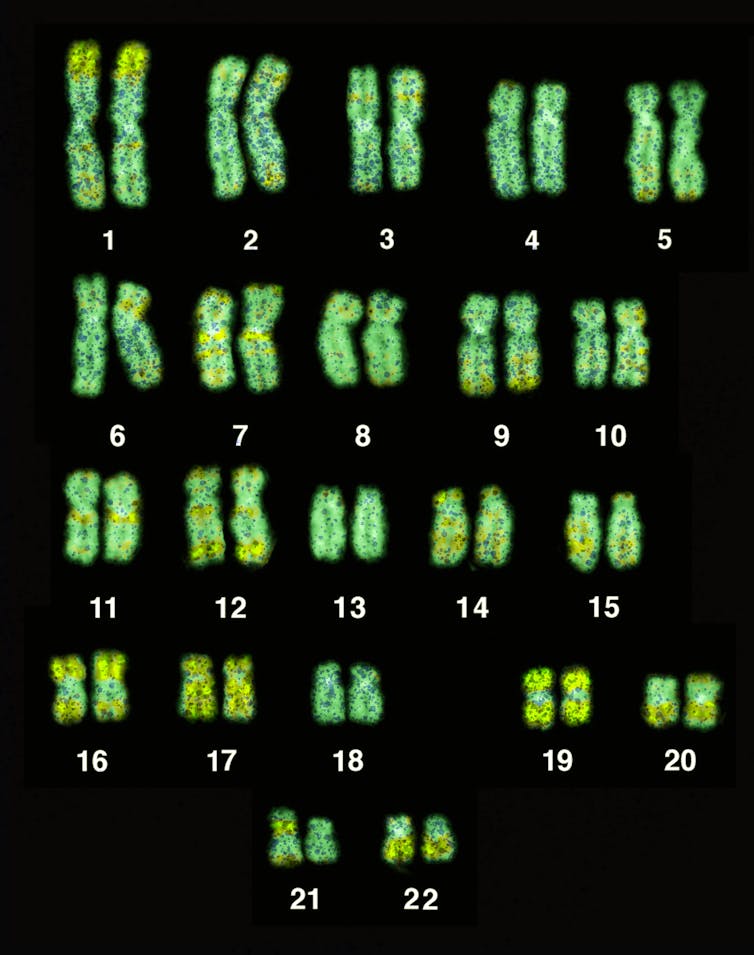 Traditionally, researchers search for individual genes that are related to disease risk by looking at tiny genetic markers all across the genome.
Andreas Bolzer, Gregor Kreth, Irina Solovei, Daniela Koehler, Kaan Saracoglu, Christine Fauth, Stefan Müller, Roland Eils, Christoph Cremer, Michael R. Speicher, Thomas Cremer, CC BY[6][7]
Traditionally, researchers search for individual genes that are related to disease risk by looking at tiny genetic markers all across the genome.
Andreas Bolzer, Gregor Kreth, Irina Solovei, Daniela Koehler, Kaan Saracoglu, Christine Fauth, Stefan Müller, Roland Eils, Christoph Cremer, Michael R. Speicher, Thomas Cremer, CC BY[6][7]
Genome-wide association studies
In 2005, researchers first used a groundbreaking method[8] called a genome–wide association study[9]. Such studies comb through huge datasets[10] of genomes and medical histories to see if people with certain diseases tend to share the same version of DNA – called a genetic marker – at specific spots.
Using this approach, researchers have identified many genes involved in Alzheimer’s disease[11]. But this method can find genetic markers only for diseases that are common in the genomes of the study participants. If, for example, 90% of participants in an Alzheimer’s disease study have European ancestry and 10% have Asian ancestry, a genome-wide association study isn’t likely to detect genetic risks for Alzheimer’s disease that are present only in individuals with Asian ancestry.
All people’s genetics reflect where their ancestors came from[12]. But ancestry manifests as both genetic variation and social and cultural experiences. All of these factors can influence risk for certain diseases, and this can create problems. When socially caused disparities in disease prevalence appear across racial groups, the genetic markers of ancestry can be mistaken for genetic markers of disease.
African Americans, for example, are up to twice as likely[13] as white Americans to develop Alzheimer’s disease[14]. Research shows that much of this disparity is likely due to structural racism[15] causing differences in nutrition, socioeconomic status and other social risk factors[16]. A genome-wide association study looking for genes associated with Alzheimer’s might mistake genetic variations associated with African descent for genetic causes of the disease.
While researchers can use a number of statistical methods[17] to avoid such mistakes, these methods can miss important findings[18] because they are often unable to overcome the overall lack of diversity in genetic datasets.
Leveraging genetics of mixed ancestry
Disentangling race, ancestry and health disparities can be a challenge in genome-wide association studies. Admixture mapping, on the other hand, is able to make better use of[19] even relatively small datasets of underrepresented people. This method specifically gets its power from studying people who have mixed ancestry.
Admixture mapping[20] relies on a quirk of human genetics – you inherit DNA in chunks[21], not in a smooth blend. So if you have ancestors from different parts of the world, your genome is made of chunks of DNA from different ancestries. This process of chunked inheritance is called admixture[22].
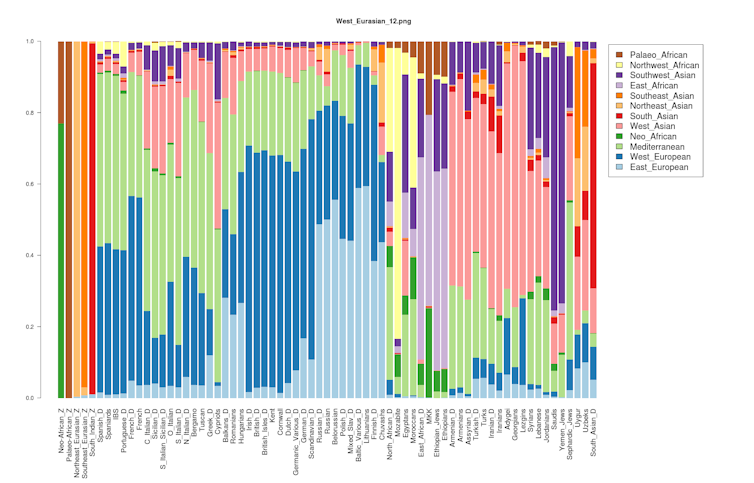 A person’s genome is made of chunks of genes that come from different ancestries.
Dodecad Project, CC BY-SA[23][24]
A person’s genome is made of chunks of genes that come from different ancestries.
Dodecad Project, CC BY-SA[23][24]
Imagine color-coding a genome by ancestry. A person who has mixed European, Native American and African ancestry might have striped chromosomes that alternate among green, blue and red, with each color representing a certain region. A different person with similar ancestry would also have a genome of green, blue and red chunks, but the order and size of the stripes would be different.
Even two biological siblings will have locations in their genomes where their DNA comes from different ancestries[25]. These ancestry stripes are how companies like Ancestry.com and 23andMe generate ancestry reports.
Because genome-wide association studies have to compare huge numbers of tiny individual genetic markers, it is much harder to find rare genetic markers for a disease. In contrast, admixture mapping tests whether the color of a certain ancestry chunk is associated with disease risk.
The statistics are fairly complicated, but essentially, because there are a smaller number of much larger ancestral chunks, it is easier to separate the signal from the noise[26]. Admixture mapping is more sensitive, but it does sacrifice specificity, as it can’t point to the individual genetic marker associated with disease risk.
Another important aspect of admixture mapping is that it looks at individuals with mixed ancestry. Since two people who have similar socioeconomic experiences can have different ancestry at certain parts of their genomes, admixture mapping can look at the association between this ancestry chunk and disease without mistaking social causes of disease for genetic causes.
Admixture mapping and Alzheimer’s disease
Researchers estimate that 58% to 79%[27] of Alzheimer’s disease risk is caused by genetic difference, but only about a third[28] of these genetic differences have been discovered. Few studies have looked for genetic links to Alzheimer’s risk among people with mixed ancestry.
Our team applied admixture mapping to a genetic dataset of Caribbean Hispanic people who have a mix of European, Native American and African ancestry. We found a part of the genome where Native American ancestry made people less likely to have Alzheimer’s disease. Essentially, we found that if you have the color blue in this certain part of your genome, you are less likely to develop Alzheimer’s disease. We believe that with further research we can find the specific gene responsible within the blue chunk and have already identified possible candidates[29].
One important note is that the genetic diversity that plays a role in disease risk is not visible to the naked eye. Anyone with Native American ancestry at this particular spot in the genome – not just a person who identifies as or looks Native American – may have some protection against Alzheimer’s disease.
Our paper illustrates that gaining a more complete understanding of Alzheimer’s disease risk requires using methods that can make better use of the limited datasets that exist for people of non-European ancestry. There is still a lot to learn about Alzheimer’s disease[30], but every new gene linked to this disease is a step toward better understanding its causes and finding potential treatments[31].
References
- ^ underrepresented in most genetic studies (gwasdiversitymonitor.com)
- ^ skewed and incomplete understanding (doi.org)
- ^ find genes (scholar.google.com)
- ^ people’s risk (scholar.google.com)
- ^ protective against Alzheimer’s disease (doi.org)
- ^ Andreas Bolzer, Gregor Kreth, Irina Solovei, Daniela Koehler, Kaan Saracoglu, Christine Fauth, Stefan Müller, Roland Eils, Christoph Cremer, Michael R. Speicher, Thomas Cremer (commons.wikimedia.org)
- ^ CC BY (creativecommons.org)
- ^ method (doi.org)
- ^ genome–wide association study (www.genome.gov)
- ^ huge datasets (doi.org)
- ^ genes involved in Alzheimer’s disease (doi.org)
- ^ reflect where their ancestors came from (doi.org)
- ^ twice as likely (www.alz.org)
- ^ develop Alzheimer’s disease (www.alz.org)
- ^ due to structural racism (dx.doi.org)
- ^ nutrition, socioeconomic status and other social risk factors (www.alz.org)
- ^ number of statistical methods (doi.org)
- ^ miss important findings (www.pbs.org)
- ^ better use of (dx.doi.org)
- ^ Admixture mapping (dx.doi.org)
- ^ inherit DNA in chunks (www.nature.com)
- ^ admixture (doi.org)
- ^ Dodecad Project (commons.wikimedia.org)
- ^ CC BY-SA (creativecommons.org)
- ^ different ancestries (www.nationalgeographic.com)
- ^ separate the signal from the noise (dx.doi.org)
- ^ 58% to 79% (doi.org)
- ^ about a third (doi.org)
- ^ identified possible candidates (doi.org)
- ^ Alzheimer’s disease (www.aarp.org)
- ^ potential treatments (dx.doi.org)
Authors: Diane Xue, Public Health Genetics PhD student and Research Assistant in Medical Genetics, University of Washington