Environmental DNA – how a tool used to detect endangered wildlife ended up helping fight the COVID-19 pandemic
- Written by Jessica Alice Farrell, PhD Candidate in Biology, University of Florida
Imagine discovering an animal species you thought had gone extinct was still living – without laying eyes on it. Such was the case with the Brazilian frog species Megaelosia bocainensis, whose complete disappearance in 1968 led scientists to believe it had become extinct. But through a novel genetic detection technique, it was rediscovered in 2020[1].
Such discoveries are now possible thanks to a new approach that recovers and reads the trace amounts of DNA released into the environment by animals. It’s called environmental DNA, or eDNA – and it takes advantage of the fact that every animal sheds DNA into its environment[2] via skin, hair, scales, feces or bodily fluids as it moves through the world.
As wildlife[3] biologists[4] at the University of Florida’s Whitney Laboratory for Marine Bioscience & Sea Turtle Hospital[5], we use eDNA to track a virus[6] responsible for a sea turtle pandemic called fibropapillomatosis[7], which causes debilitating tumors[8]. We also use eDNA to detect sea turtles in the wild.
But in 2020, human health researchers began repurposing eDNA techniques to track the COVID-19 pandemic. This is a prime example of how research in one area – wildlife conservation – can be adapted to another area – human disease mitigation. Going forward, we believe eDNA will prove to be an essential tool for monitoring both human and animal health.
From soil microbes to sea turtles
Scientists in the 1980s began hunting for microbe DNA in soil samples[9]. Over the next 20 years, the technique was adapted for use with air and water samples, and scientists started using eDNA to detect larger animals[10] and plants[11].
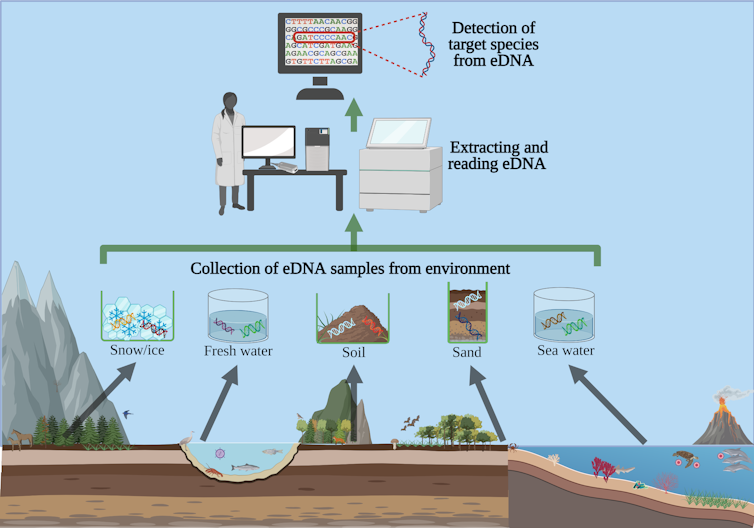 Scientists now can detect DNA traces from many different environments.
Liam Whitmore, University of Limerick, CC BY-ND[12]
Scientists now can detect DNA traces from many different environments.
Liam Whitmore, University of Limerick, CC BY-ND[12]
While the science behind eDNA techniques is complex, the actual process of collecting and testing a sample is relatively simple. Samples are filtered through very fine paper, which traps loose cells and strands of DNA. The techniques to read what DNA is present are the same as those used for tissue or blood samples, usually quantitative polymerase chain reaction[13] or whole genome sequencing[14]. Scientists can either read all of the DNA present from every organism – or target just the DNA from species of interest.
Scientists now routinely use eDNA to detect endangered wildlife and invasive species[15]. The ability to tell whether an animal is present without ever needing to lay eyes or a lens on it is an incredible leap forward, decreasing the time, resources and human effort needed to monitor and protect vulnerable species.
 Routine imaging of a juvenile green sea turtle patient afflicted with virus-triggered fibropapillomatosis at the Florida Whitney Sea Turtle Hospital.
Devon Rollinson-Ramia, CC BY-ND[16]
Routine imaging of a juvenile green sea turtle patient afflicted with virus-triggered fibropapillomatosis at the Florida Whitney Sea Turtle Hospital.
Devon Rollinson-Ramia, CC BY-ND[16]
However, to truly protect endangered species, it’s not just the animals that need to be monitored, but the pathogens that threaten their survival. Environmental DNA is able to monitor the parasites, fungi and viruses that can cause disease in wildlife[17].
Tracking COVID-19
While scientists originally applied eDNA to human pathogen detection[18] over a decade ago, it wasn’t until the beginning of the current COVID-19 human pandemic that the repurposing of eDNA took off on a large scale, allowing the technology to make staggering advancements in very short order.
Coronavirus genomes consist not of DNA, but rather its cousin molecule, RNA. So researchers have rapidly optimized a variation of eDNA – eRNA – to detect coronavirus RNA in air and human wastewater.
For example, at the University of Florida Health Shands Hospital, researchers collected air samples from the hospital room of two COVID-19 patients. Using eRNA, they successfully isolated and sequenced the virus[19]. Confirming air as a key route of transmission directly influenced public health guidelines.
 Collecting sewage samples to test for SARS-CoV-2 at Utah State University in September 2020.
AP Photo/Rick Bowmer[20]
Collecting sewage samples to test for SARS-CoV-2 at Utah State University in September 2020.
AP Photo/Rick Bowmer[20]
When scientists apply eRNA to archived wastewater samples[21], the true dates of SARS-CoV-2 appearance can be detected[22]. SARS-CoV-2 concentration in wastewater in Valencia, Spain[23], peaked on March 9, 2020, but the number of clinical cases didn’t peak until the start of April 2020 because of the lag time between infection and severe clinical symptoms.
This sort of predictive monitoring has profound implications for health care systems, allowing time to prepare – not just for COVID-19, but for any future disease outbreaks that threaten human populations.
[Understand new developments in science, health and technology, each week. Subscribe to The Conversation’s science newsletter[24].]
Intersection of diseases
It’s vital that human and animal diseases are studied together. Sixty percent of emerging human pathogens come from animals[25] – with many of these (42%) originating in wildlife populations, including Ebola, Zika, West Nile and Marburg viruses. Alternatively, people can also transmit pathogens to animals[26].
SARS-CoV-2 has already infected apes at a zoo in San Diego[27], large cats at a zoo in New York[28] and minks at farms in Europe[29] - the latter of which gave rise to new variants that could prove a new threat to people[30].
Medics, veterinarians and scientists call this convergence of human, animal and environmental well-being OneHealth[31] or EcoHealth. Studying and treating human and wildlife disease together recognizes their commonalities[32] and often yields breakthroughs.
With eDNA, all pathogens can be monitored in an environment regardless of where they come from. An integrated eDNA monitoring program could cost-effectively provide advanced warning of human, livestock and wildlife diseases.
References
- ^ it was rediscovered in 2020 (doi.org)
- ^ every animal sheds DNA into its environment (doi.org)
- ^ wildlife (scholar.google.com)
- ^ biologists (scholar.google.com)
- ^ Whitney Laboratory for Marine Bioscience & Sea Turtle Hospital (www.whitney.ufl.edu)
- ^ use eDNA to track a virus (doi.org)
- ^ sea turtle pandemic called fibropapillomatosis (doi.org)
- ^ causes debilitating tumors (theconversation.com)
- ^ hunting for microbe DNA in soil samples (doi.org)
- ^ detect larger animals (doi.org)
- ^ and plants (doi.org)
- ^ CC BY-ND (creativecommons.org)
- ^ quantitative polymerase chain reaction (www.theguardian.com)
- ^ whole genome sequencing (www.theguardian.com)
- ^ detect endangered wildlife and invasive species (doi.org)
- ^ CC BY-ND (creativecommons.org)
- ^ viruses that can cause disease in wildlife (doi.org)
- ^ applied eDNA to human pathogen detection (doi.org)
- ^ successfully isolated and sequenced the virus (doi.org)
- ^ AP Photo/Rick Bowmer (newsroom.ap.org)
- ^ archived wastewater samples (doi.org)
- ^ true dates of SARS-CoV-2 appearance can be detected (doi.org)
- ^ SARS-CoV-2 concentration in wastewater in Valencia, Spain (doi.org)
- ^ Subscribe to The Conversation’s science newsletter (theconversation.com)
- ^ Sixty percent of emerging human pathogens come from animals (www.rte.ie)
- ^ people can also transmit pathogens to animals (science.thewire.in)
- ^ apes at a zoo in San Diego (apnews.com)
- ^ large cats at a zoo in New York (www.nationalgeographic.com)
- ^ minks at farms in Europe (www.theguardian.com)
- ^ gave rise to new variants that could prove a new threat to people (www.who.int)
- ^ OneHealth (www.cdc.gov)
- ^ recognizes their commonalities (www.zoobiquity.com)
Authors: Jessica Alice Farrell, PhD Candidate in Biology, University of Florida