A celebrated AI has learned a new trick: How to do chemistry
- Written by Marc Zimmer, Professor of Chemistry, Connecticut College
Artificial intelligence has changed the way science is done by allowing researchers to analyze the massive amounts of data modern scientific instruments generate. It can find a needle in a million haystacks of information and, using deep learning[1], it can learn from the data itself. AI is accelerating advances in gene hunting[2], medicine[3], drug design[4] and the creation of organic compounds[5].
Deep learning uses algorithms, often neural networks that are trained on large amounts of data, to extract information from new data. It is very different from traditional computing with its step-by-step instructions. Rather, it learns from data. Deep learning is far less transparent than traditional computer programming, leaving important questions – what has the system learned, what does it know?
As a chemistry professor[6] I like to design tests that have at least one difficult question that stretches the students’ knowledge to establish whether they can combine different ideas and synthesize new ideas and concepts. We have devised such a question for the poster child of AI advocates, AlphaFold, which has solved the protein-folding problem[7].
Protein folding
Proteins are present in all living organisms. They provide the cells with structure, catalyze reactions, transport small molecules, digest food and do much more. They are made up of long chains of amino acids like beads on a string. But for a protein to do its job in the cell, it must twist and bend into a complex three-dimensional structure, a process called protein folding. Misfolded proteins can lead to disease.
In his chemistry Nobel acceptance speech in 1972, Christiaan Anfinsen[8] postulated that it should be possible to calculate the three-dimensional structure of a protein from the sequence of its building blocks[9], the amino acids.
Just as the order and spacing of the letters in this article give it sense and message, so the order of the amino acids determines the protein’s identity and shape, which results in its function.
Because of the inherent flexibility of the amino acid building blocks, a typical protein can adopt an estimated 10 to the power of 300 different forms[11]. This is a massive number, more than the number of atoms in the universe[12]. Yet within a millisecond every protein in an organism will fold into its very own specific shape – the lowest-energy arrangement of all the chemical bonds that make up the protein. Change just one amino acid in the hundreds of amino acids typically found in a protein and it may misfold and no longer work.
AlphaFold
For 50 years computer scientists have tried to solve the protein-folding problem – with little success. Then in 2016 DeepMind[13], an AI subsidiary of Google parent Alphabet, initiated its AlphaFold[14] program. It used the protein databank[15] as its training set, which contains the experimentally determined structures of over 150,000 proteins.
In less than five years AlphaFold had the protein-folding problem beat[16] – at least the most useful part of it, namely, determining the protein structure from its amino acid sequence. AlphaFold does not explain how the proteins fold so quickly and accurately. It was a major win for AI, because it not only accrued huge scientific prestige, it also was a major scientific advance that could affect everyone’s lives.
Today, thanks to programs like AlphaFold2[17] and RoseTTAFold[18], researchers like me can determine the three-dimensional structure of proteins from the sequence of amino acids that make up the protein – at no cost – in an hour or two. Before AlphaFold2 we had to crystallize the proteins and solve the structures using X-ray crystallography[19], a process that took months and cost tens of thousands of dollars per structure.
We now also have access to the AlphaFold Protein Structure Database[20], where Deepmind has deposited the 3D structures of nearly all the proteins found in humans, mice and more than 20 other species. To date they it has solved more than a million structures and plan to add another 100 million structures this year alone. Knowledge of proteins has skyrocketed. The structure of half of all known proteins is likely to be documented by the end of 2022, among them many new unique structures associated with new useful functions.
Thinking like a chemist
AlphaFold2 was not designed to predict how proteins would interact with one another, yet it has been able to model how individual proteins combine to form large complex units composed of multiple proteins[21]. We had a challenging question for AlphaFold – had its structural training set taught it some chemistry? Could it tell whether amino acids would react with one another – a rare yet important occurrence?
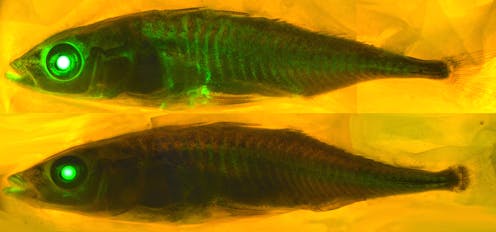
I am a computational chemist interested in fluorescent proteins[22]. These are proteins found in hundreds of marine organisms like jellyfish and coral. Their glow can be used to illuminate[23] and study diseases[24].
There are 578 fluorescent proteins in the protein databank[26], of which 10 are “broken” and don’t fluoresce. Proteins rarely attack themselves, a process called autocatalytic posttranslation modification, and it is very difficult to predict which proteins will react with themselves and which ones won’t.
Only a chemist with a significant amount of fluorescent protein knowledge would be able to use the amino acid sequence to find the fluorescent proteins that have the right amino acid sequence to undergo the chemical transformations required to make them fluorescent. When we presented AlphaFold2 with the sequences of 44 fluorescent proteins that are not in the protein databank, it folded the fixed fluorescent proteins differently from the broken ones[27].
The result stunned us: AlphaFold2 had learned some chemistry. It had figured out which amino acids in fluorescent proteins do the chemistry that makes them glow. We suspect that the protein databank training set and multiple sequence alignments[29] enable AlphaFold2 to “think” like chemists and look for the amino acids required to react with one another to make the protein fluorescent.
A folding program learning some chemistry from its training set also has wider implications. By asking the right questions, what else can be gained from other deep learning algorithms? Could facial recognition algorithms find hidden markers for diseases? Could algorithms designed to predict spending patterns among consumers also find a propensity for minor theft or deception? And most important, is this capability – and similar leaps in ability[30] in other AI systems – desirable?
References
- ^ deep learning (www.techtarget.com)
- ^ gene hunting (doi.org)
- ^ medicine (doi.org)
- ^ drug design (news.mit.edu)
- ^ the creation of organic compounds (doi.org)
- ^ chemistry professor (scholar.google.ca)
- ^ protein-folding problem (doi.org)
- ^ Christiaan Anfinsen (www.nobelprize.org)
- ^ calculate the three-dimensional structure of a protein from the sequence of its building blocks (directorsblog.nih.gov)
- ^ CC BY-ND (creativecommons.org)
- ^ 10 to the power of 300 different forms (web.archive.org)
- ^ number of atoms in the universe (educationblog.oup.com)
- ^ DeepMind (www.deepmind.com)
- ^ AlphaFold (www.deepmind.com)
- ^ protein databank (www.rcsb.org)
- ^ the protein-folding problem beat (www.deepmind.com)
- ^ AlphaFold2 (www.deepmind.com)
- ^ RoseTTAFold (www.ipd.uw.edu)
- ^ X-ray crystallography (doi.org)
- ^ AlphaFold Protein Structure Database (alphafold.ebi.ac.uk)
- ^ form large complex units composed of multiple proteins (www.nature.com)
- ^ fluorescent proteins (theconversation.com)
- ^ to illuminate (theconversation.com)
- ^ study diseases (global.oup.com)
- ^ Wen Lu and Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University (images.nigms.nih.gov)
- ^ protein databank (www.rcsb.org)
- ^ it folded the fixed fluorescent proteins differently from the broken ones (journals.plos.org)
- ^ CC BY-ND (creativecommons.org)
- ^ multiple sequence alignments (samfordubioinformatics.wordpress.com)
- ^ similar leaps in ability (www.technologyreview.com)
Authors: Marc Zimmer, Professor of Chemistry, Connecticut College
Read more https://theconversation.com/a-celebrated-ai-has-learned-a-new-trick-how-to-do-chemistry-182031