Where coronavirus variants emerge, surges follow – new research suggests how genomic surveillance can be an early warning system
- Written by Bart C. Weimer, Professor of Population Health & Reproduction, University of California, Davis
Genomic surveillance programs have let scientists track the coronavirus over the course of the pandemic. By testing patient samples, researchers are able to diagnose COVID-19. But they’re also able to use genetic changes in the virus to recreate its travel routes and identify the emergence of new viral variants.
As microbiologists[1], we examined how quickly the coronavirus genome has mutated during the pandemic and then figured out how quickly these changes led to new cases[2] and rapid disease spread.
By connecting genetic change with the appearance of new clusters of disease, our research suggests how genome surveillance can provide a new early warning of what’s to come. Daily reports on how the virus is evolving could sound the alarm before case numbers explode.
Mutations happen and can be tracked
Starting around 2012, researchers began to develop genome sequencing as a way for public health experts to track infectious diseases[3]. Basically they are able to “read” an organism’s whole genetic code, the long list of A, C, G and T molecules that comprise the blueprints for the proteins that carry out the cell’s functions.
When pathogens infect a host, they reproduce themselves. Changes to the genetic code can happen at this point – like typos you might make copying down a page of text, substituting an A for a T in one spot, for instance. These changes are mutations. They provide new instructions to the next generation that can give them new capabilities – maybe they are better able to move between hosts, survive and initiate outbreaks or cause new symptoms.
Multiple versions of the same organism, but with variations in the genetic code, circulate during a disease outbreak. Depending on how successful they are at infecting new hosts and spreading, various versions can become more or less common.
Historically, public health labs tracked disease outbreaks by the name of the pathogen – SARS, salmonella, Ebola and so on. But as the speed and accuracy of genome sequencing increased, researchers realized that the same pathogen can be divided into many different subpopulations based on genetic variation.
These are the variants you hear about with regard to the coronavirus – the B.1.1.7 strain that first emerged in the U.K., the B.1.617 version that was identified in India, and the B.1.427 and B.1.429 variants that both originated in California. All are technically classified as the same SARS-CoV-2 virus, but they may have quite different features.
Screening isn’t the same as sequencing
When a person’s sample is tested for SARS-CoV-2, the lab uses a technique called PCR[4] to identify whether certain coronavirus genes are present. This method is good for screening – diagnosing whether the person in fact has COVID-19 or not. It also provides important surveillance data about how many people have the coronavirus in a particular time and place.
But it doesn’t sequence the whole genome, which is made up of 30,000 nucleotides – those As, Gs, Cs and Ts. The PCR screening test just looks for one small stretch of the coronavirus’s genetic code – the gene related to the virus’s spike protein that helps it infect human cells. This technique won’t flag mutations happening in other parts of the genome because it’s not looking for them.
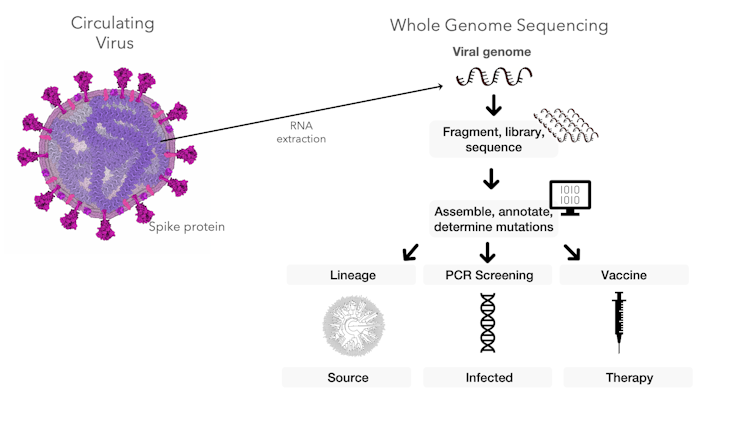 Sequencing the genetic material of the coronavirus can help researchers trace the travel routes of the virus, diagnose infected people and inform research into vaccines and therapeutics.
Bart Weimer and Darwin Bandoy, CC BY-ND[5]
Sequencing the genetic material of the coronavirus can help researchers trace the travel routes of the virus, diagnose infected people and inform research into vaccines and therapeutics.
Bart Weimer and Darwin Bandoy, CC BY-ND[5]
Other mutations are definitely occurring, though. Sequencing the entire genomes of coronavirus samples creates a massive list of variants. Our work tackles this ever-changing list to show that not only do mutations in the spike gene lead to new outbreak clusters – additional mutations in other genes increase outbreaks, too.
Connecting variants and outbreaks
To figure out the role of these mutations, we directly linked the variants present at a certain time and place with the coronavirus’s reproductive number, known as R for short[6]. R is a way to quantify the intensity of an infectious disease outbreak. It stands for how many additional people an infected person will spread the germ to.
But R doesn’t tell you what version of the viral genome was passed along. By directly linking R and the variant present, we were able to pinpoint the specific mutation that was emerging and increasing viral spread. We found that as new variants became more common, COVID-19 diagnoses surged.
By merging genomics with classical epidemiology, we created a tool that factors in rising variants and R to warn how quickly cases will spread and which variants are more likely to trigger new outbreaks.
To test this approach, we linked the SARS-CoV-2 genotype to the daily R during the first three months of the pandemic using 150 genomes. Our method predicted the near future of outbreaks in four different countries that each had various levels of mandated social interventions.
This preliminary evidence relied on a small number of genome sequences, but it was all the data available from the early stages of the pandemic. As the pandemic continues, labs are sequencing thousands of genomes[7] across the globe weekly. We replicated our initial estimates using 20,000 genomes from the U.K. and arrived at the same observation – new variants led to more transmission, variants are continuing to expand and will continue to increase in prevalence as the pandemic continues.
By incorporating genome sequencing data with information about transmissibility, we created a kind of early warning system, allowing us to forecast spreading events. In the real world, advance warning like this could inform public health decisions about social interventions. People can prepare for predicted outbreaks. A bonus is that our model also would show when highly contagious variants are declining – providing solid evidence to support loosening restrictions to allow a return to normalcy.
 Just as valuable as early warning, variant information could help officials know when it’s safer to lift restrictions.
SOPA Images/LightRocket via Getty Images[8]
Just as valuable as early warning, variant information could help officials know when it’s safer to lift restrictions.
SOPA Images/LightRocket via Getty Images[8]
Scanning the horizon for future threats
We believe that public health is at the dawn of integrating genome sequencing with infectious disease tracking. We envision a reference library of pathogen genomes, representing the diversity of their many emerging variants. It could be a new tool for epidemiologists, a part of routine surveillance programs that can last beyond the current pandemic.
In the future, scientists hopefully won’t need to wait for an outbreak to grow. Our research suggests that by identifying a rise in variants early, public health officials can quickly respond – before the inevitable rise in new disease cases. We think this kind of early warning system can increase the public’s safety for any pathogen and reduce outbreaks for all types of organisms.
[Over 100,000 readers rely on The Conversation’s newsletter to understand the world. Sign up today[9].]
References
- ^ microbiologists (scholar.google.com)
- ^ figured out how quickly these changes led to new cases (doi.org)
- ^ to track infectious diseases (doi.org)
- ^ a technique called PCR (my.clevelandclinic.org)
- ^ CC BY-ND (creativecommons.org)
- ^ reproductive number, known as R for short (theconversation.com)
- ^ labs are sequencing thousands of genomes (doi.org)
- ^ SOPA Images/LightRocket via Getty Images (www.gettyimages.com)
- ^ Sign up today (theconversation.com)
Authors: Bart C. Weimer, Professor of Population Health & Reproduction, University of California, Davis